GPU accelerated image processing of Cryo-EM data determining a high-resolution structure of the Thermosome a molecular machine
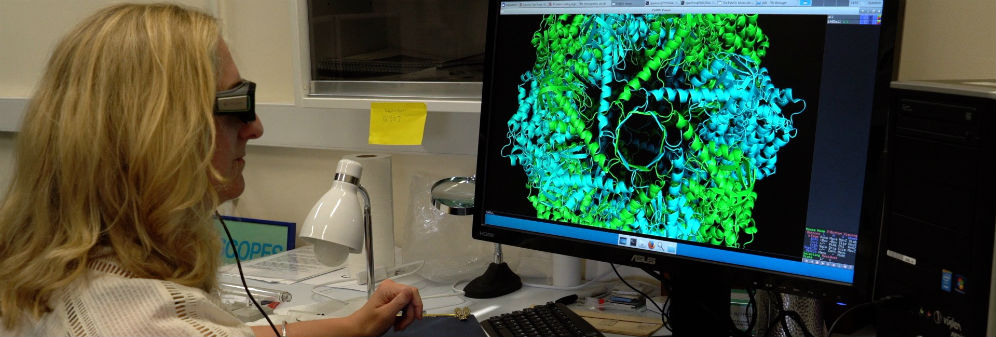
With a trip to the National Electron Microscopy (EM) facility and the expertise of a Junior Research Software Engineer, this project built a GPU machine and cooling system
Specialist input from the Research Computing team
There was a requirement to advance the compute intensive projects in Dr Paul's lab and the desire to have specialist input from the Research Computing team.
The project team set up and installed specialist software to enable the high-resolution image processing of the 3D structure of the Thermosome, a class ll chaperonin, captured using Cryo-Electron microscopy and single particle image processing. Specifically, to facilitate the analysis of data collected at the national facility of EM in Harwell, and to determine a high-resolution 3D structure of the Thermosome, a molecular machine. Due to the quantity of data generated (4-8TB) and the specific software that is used, written for GPU processing, a new dedicated machine was required.
Further funding bids and benefits to other faculties
The data generated with this machine will form pilot data for future grant proposals including a planned project studying the ultra-structure of the thermosome to the BBSRC research council. Projects from many different departments will also benefit from this machine, bringing together different faculties across the University of Bristol.
Collaboration
The seed corn funding developed a collaboration between Dr Danielle Paul from the School of Physiology, Pharmacology and Neuroscience and Dr Matt Williams from the Advanced Centre for Research Computing (ACRC) to advance the compute intensive projects in Dr Paul's lab.
People involved in this project
- Dr Danielle Paul (BHF Research Fellow)
- Dr Matt Williams (Junior Research Software Engineer)