Stage III Project - Grant Elmes
Expresion and Analysis of Polyketide Synthase Genes
Stage III Project - Grant Elmes
SGM Summer Student - Rosie Stentiford
Supervisor Dr Russell Cox
Second Assesor Professor T. J. Simpson
Background
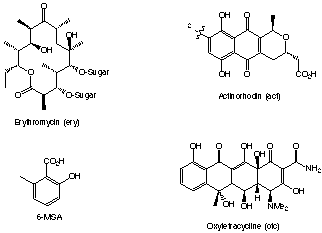
Polyketides are a class of naturally occurring molecules which often
have important biological properties. For example oxytetracycline and erythromycicn
are important commercial antibiotics. Generally theses compounds are too
complex for total synthesis to be a viable way of obtaining large quantities
for commercial exploitation. Fermentation of bacterial or fungal strains
followed by extraction and purification are preferered by pharmaceutical
companies.
Genes coding for polyketide production have been found in many fungal
and bacterial species. However it is difficult to confirm that a particular
set of genes is involved in the production of a particular polyketide because
many organisms posess more than a single set of polyketide genes. In order
to identify what a particular polyketide synthase gene produces this project
will investigate the possibility of inserting individual genes into a bacterium
which is known to produce no polyketides (Streptomyces coelicolor
CH999). The genes are expected to be translated into proteins in the usual
way and the proteins should then synthesise the relevent polyketide. Extraction
and identification of the polyketide will then be performed. This is the
first step in an important long-term effort to produce novel antibiotics
through genetic modification of PKS genes.
Programme
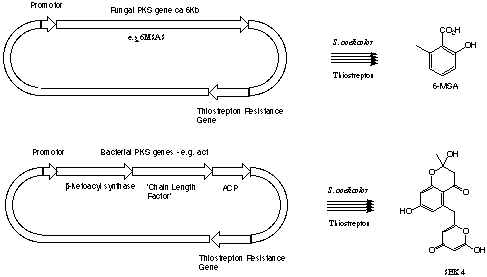
Streptomyces species have been shown recently to be suitable hosts
for the expression of heterologous genes (e.g from fungi and other bacteria).
In particular, the expression of recombinant fungal polyketide synthase
(PKS) genes has been demonstrated. PKS enzymes require post-translational
modification in order to affix a 4'-phosphopantetheine prosthetic group
to the acyl carrier protein sub-domain, catalysed by so-called holo-ACP
synthase. Expression of 6-methyl salicylic acid synthase (6MSAS) by S.
coelicolor CH999 leads to high production levels of the metabolite,
indicating correct post translational modification by this bacterium.
This project will assess expression of a recombinant putative norsolorinic
acid synthase gene from the fungus Aspergillus parasiticus and putative
minimal actinorhodin genes from the bacteria Streptomyces coelicolor.
The bacteria S. coelicolor CH999 which produces no polyketides will
be used as the host organism (S. lividans has been used successfully
for 6MSAS in this laboratory but results are confused by background natural
polyketide production). Standard microbiological methods to grow a number
of strains of S. coelicolor CH999 containing expression vectors
for the putative PKS genes, as well as similar control strains carrying
a 6MSA expression vector. Extraction of the culture broths will be followed
by HPLC and NMR analysis. Synthetic standards of 6MSA, norsolorinic acid
and SEK4/4b (produced by the act genes) are available for comparative purposes.
The relevent genetic constructs have been synthesised and are ready for
expression.
References
. D. J. Bedford, E. Schweizer, D. A. Hopwood and C. Khosla, J. Bacteriol.,
1995, 177, 4544-4548.
. R. J. Cox, T. S. Hitchman, K. J. Byrom, I. S. C. Findlow, J. A. Tanner,
J. Crosby and T. J. Simpson, FEBS Letters, 1997, 405, 267-272.
. R. A. Stentiford, Summmer Project 1997.
. Fungal PKS by Dr Colin Lazarus Dept. Botany, bacterial PKS by Dr Anne-Lise
Matharu and Dr R. J. Cox, School of Chemistry.