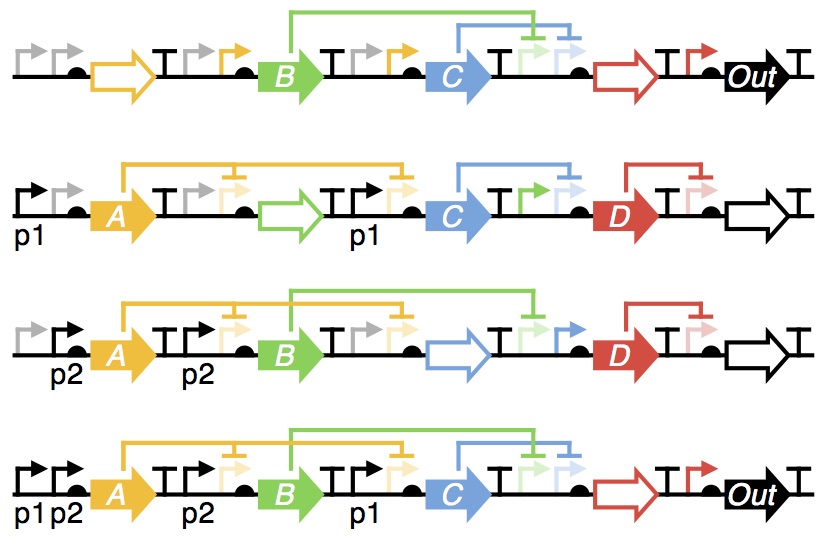
Synthetic genetic circuits
Synthetic genetic circuits allow for the reprogramming of cellular functions and form the foundation of many transformative applications in synthetic biology. Nevertheless, after more than a decade of development, the rational design of their function remains elusive. This stems from our limited under-standing of how genetic parts behave when combined in new ways and an inability to observe the behaviour of every component when failures do arise.
This project aims to apply experimental and theoretical methods to support the rational design of large synthetic genetic circuits and significantly reduce the time to create functional systems. Methods such as deep-sequencing, genome-scale models and in vitro platforms are being used in conjunction with carefully designed libraries of genetic circuits, in which the order, orientation and strength of components are varied. These libraries explore the various contexts that genetic parts may face and measurements of their performance using our high-throughput and quantitative methodologies will help reveal the uncharacterised interactions that can occur. From these measurements, we hope to extract "engineering rules" that capture those aspects of a design critical for reliable performance and incorporate these into new circuit design processes. This project will provide synthetic biologists with an unprecedented window into the workings of their systems and help capture and predict unwanted interactions between the components and the host cell.
Project lead: BrisSynBio Fellow in Synthetic Biology Dr Thomas Gorochowski