MULTICOLI
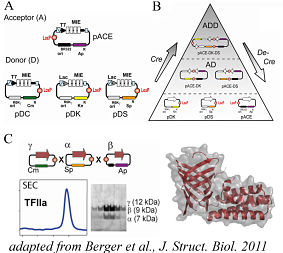
To date, E.coli remains the major work-horse for recombinant protein production in most research laboratories, for many good reasons. The vast majority of entries in the Protein Data Bank (PDB) constitute amino acids chains that were recombinantly produced in E.coli. Numerous complexes have been successfully produced using this prokaryotic host, in the quality and quantity required for high-resolution structural studies. Several co-expression systems exist that facilitate the heterologous production of proteins and their complexes inE.coli. The MultiColi system applies tandem recombineering (TR) for multiprotein complex production in this host, optionally in robot-assisted high-throughput mode for structural genomics-type applications.
MultiColi uses an array of Donors and Acceptors that are charged with one or several protein encoding genes, in form of individual expression cassettes and/or polycistrones. Polycistrones are concatenated genes, spaced apart by ribosome binding sites and transcriptional enhancers, under the control of a single promoter. The tandem recombineering (TR) method was developed by the Berger laboratory using E.coli as a model host system, and implemented in the first fully roboterised pipeline (ACEMBL) for multiprotein complex production. The protocols that were scripted into automated procedures were then used to adapt MultiBac and MultiMam as eukaryotic multiexpression derivatives to the ACEMBL HT pipeline.
For in silico generating Donor-Acceptor fusion constructs you can use Cre-ACEMBLER.
For further information on MultiColi contact imre.berger@bristol.ac.uk